Interactive Dynamics
Integrating the Dynamics of the Ribosomal Allocation Model
As presented in the main text of the paper, a simple model of microbial growth focused on the accumulation of biomass can be summarized by the following handful of differential equations:
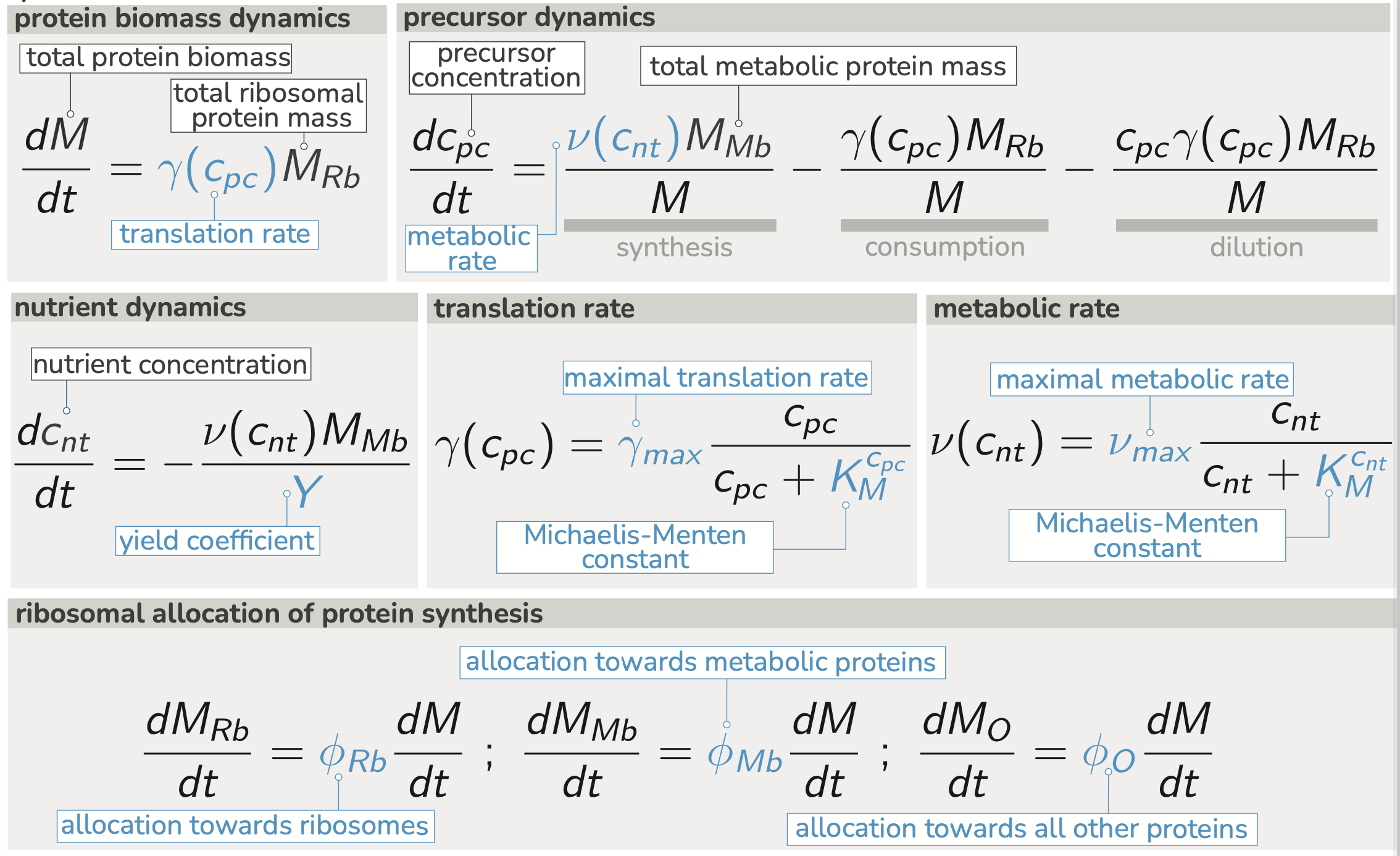
We can numerically integrate these equations to get a sense for how the model operates. In Figure 2 of the main text, we show the integrated dymamics of the biomass accumulation, the precursor concentration, and the nutrient concentration of the system for a single set of parameter values. To gain a better intuition for how the model works (and how important the relevant parameters are), it is helpful to interactively change them and observe the result. The interactive figure below allows just that.
Note that this integration uses a simple forward-Euler method and thus can be unstable in some parameter regimes, particularly at the extreme values permitted via the sliders.