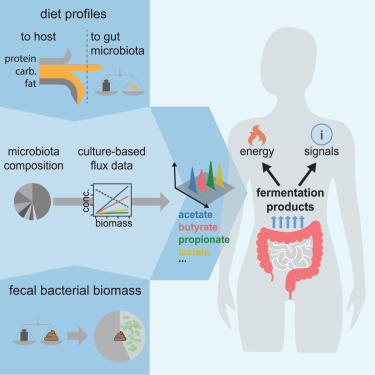
Fermentation Products Analysis
Code, data, and analysis quantifying the varying harvest of fermentation products from the human gut microbiota across dietary conditions.
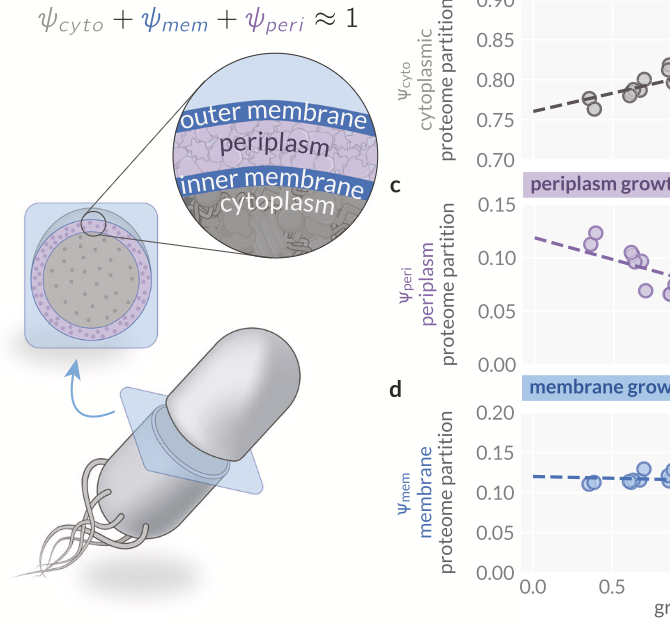
Envelope Proteome and the Density Maintenance Model
Code and analysis for the study of cytoplasmic and membrane density maintenance and its role in shaping cellular geometry in E. coli.
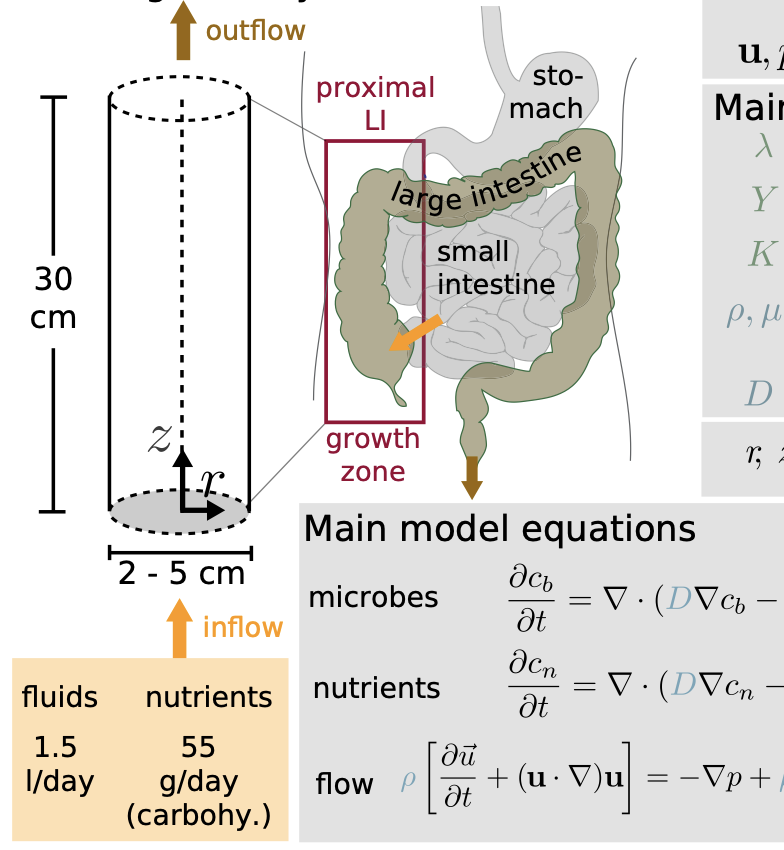
Diurnal Growth Variation Modeling
Modeling and analysis of diurnal variations in gut microbial growth dynamics driven by host feeding rhythms.
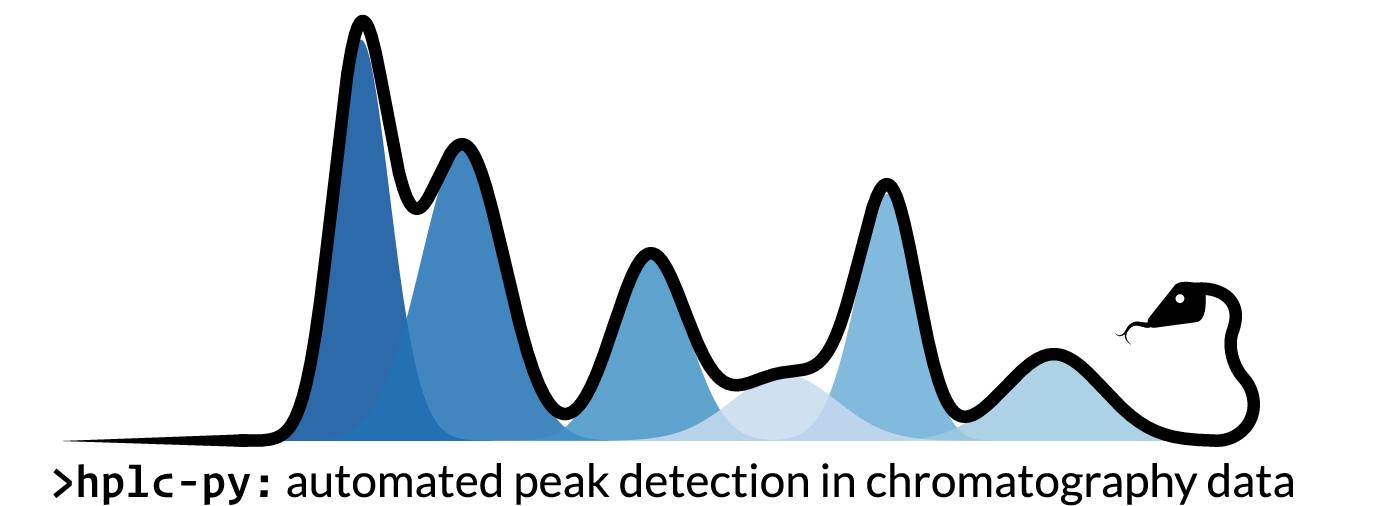
hplc-py
A Python package for rapid, automated peak quantification in complex chromatograms. Designed for high-throughput analysis of HPLC data in microbial physiology experiments.
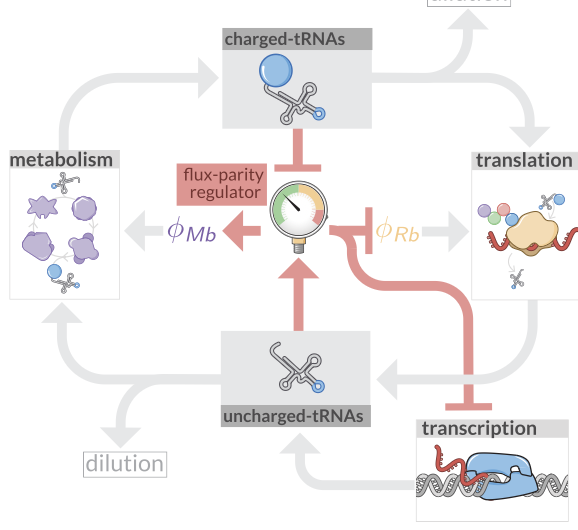
Optimal Allocation and Flux Parity Model
Code and analysis for the flux parity model of optimal resource allocation in microbial growth.
Collective Movement by Chemotaxis
Simulations and analysis of collective bacterial range expansion driven by chemotaxis.
Analyzing Bacterial Swimming Behavior
Tools for quantifying individual bacterial swimming trajectories and run-tumble statistics from microscopy data.
Modeling Growth of Human Gut Microbiota
Mathematical models linking bacterial physiology, flow, and nutrient gradients to microbiota composition in the human gut.